Hot Products
Top 100 Antibodies
Customer Testimonials
"I was having some issues with Phire Direct PCR Kits. I tried out your LiDirect™ Lightning Genotyping Kit and it worked great! Thanks y'all!"
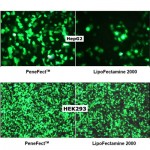
"We are happy to share with you that the sample given to us has worked very good. The results are really satisfactory. We would like to shift totally towards LiFect293 transfection reagent from Lipofectamine and Fugene for our experiments in future."- - - - - - - - - - - - - - - - - - - - - - -