VAHTS Universal DNA Library Prep Kit for Illumina V4 (24 rxns)
VAHTS Universal DNA Library Prep Kit for Illumina V4 is a library preparation kit targeted for Illumina high-throughput sequencing platform optimization. 100 pg -1 μg of fragmented double-stranded DNA can be converted into a dedicated library for Illumina high-throughput sequencing platform. As a new generation of upgraded version, the kit can improve the conversion rate of low-quality template libraries and reduce the repetition rate of libraries by optimizing and improving the terminal repair module, connecting module and library amplification module. This kit is widely suitable for PCR or PCR-Free library preparation of a variety of samples, and is compatible with the targeted capture process. All reagents provided in the kit have undergone strict quality control and functional validation to ensure the stability and repeatability of library preparation to the greatest extent.
Key Features
• Wide Compatibility: Compatible with FFPE and cfDNA samples of low quality in addition to conventional samples.
• High Efficiency: Higher library conversion rate.
• Accurate Detection: Accurate detection of low-frequency mutations.
Applications
1. Whole genome sequencing
2. Whole-exome sequencing or other targeted capture sequencing
3. Amplicon sequencing
4. ChIP-seq
5. Metagenome sequencing
6. Methylation Sequencing
Components
1. End Prep Buffer 2: 168 µl
2. End Prep Enzyme 2: 72 µl
3. Rapid Ligation Buffer 5: 720 μl
4. Rapid DNA Ligase 2: 240 μl
5. VAHTS HiFi Amplification Mix 2: 600 μl
6. PCR Primer Mix 3 for Illumina: 120 μl
7. Control DNA (264 bp, 50 ng/μl): 10 μl
Storage
Store at -20°C.
Case Study
1. Suitable for low-quality FFPE, cfDNA, etc.
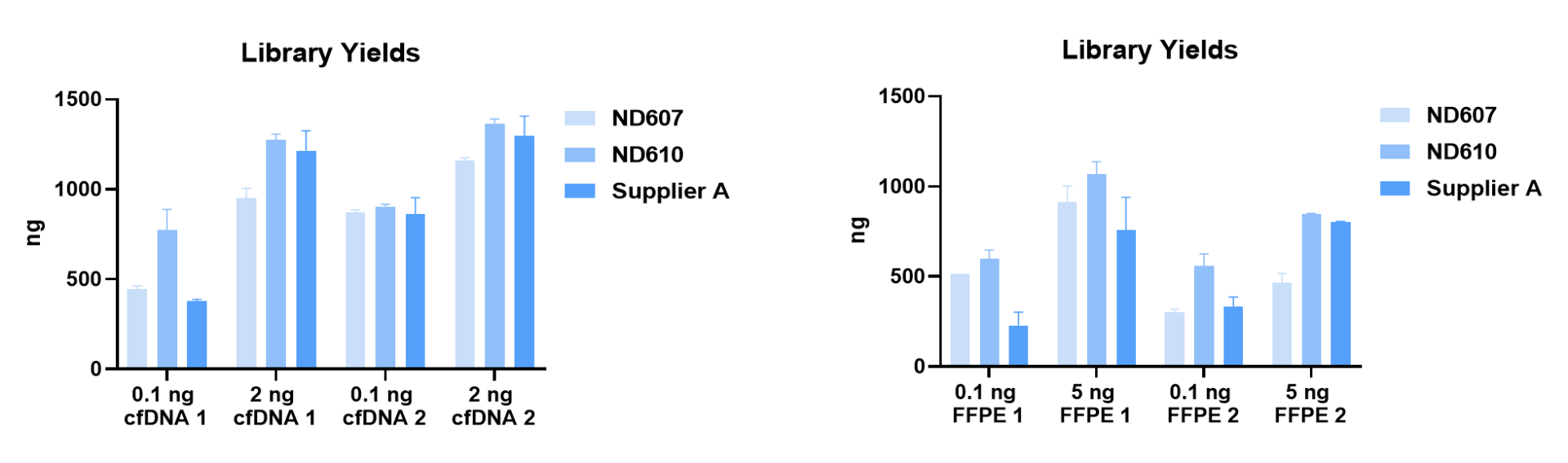
Different input amounts of clinical FFPE and cfDNA were used as templates for library construction. Under the same number of amplification cycles, the ND610 library output had significant advantages over similar products of Supplier A.
2. Higher library conversion rate
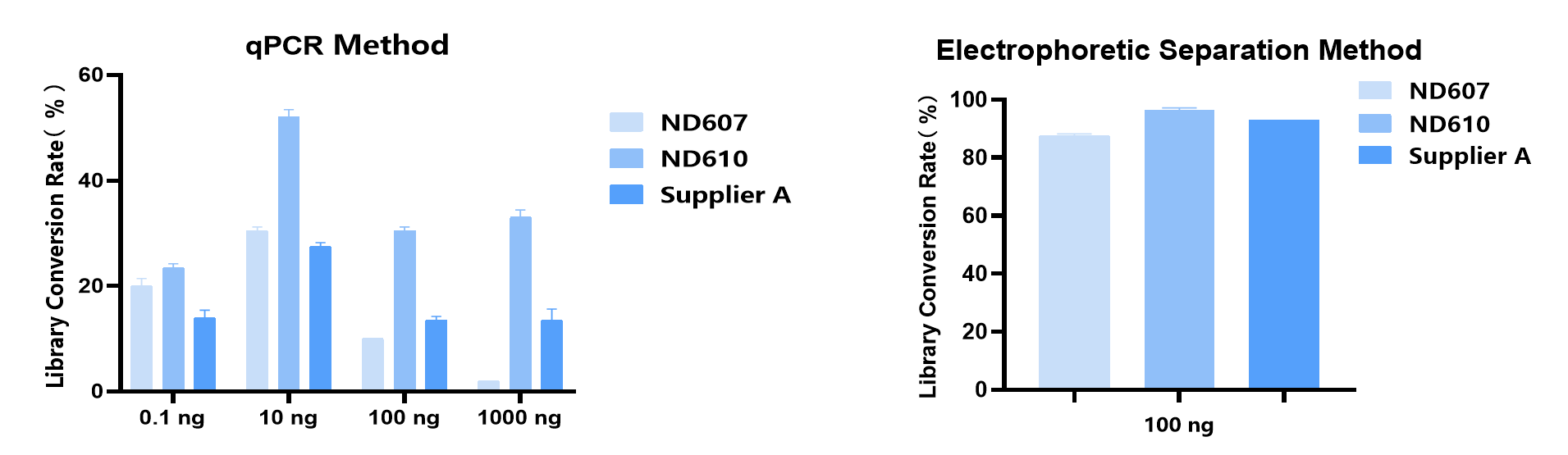
Different samples and different input amounts were used for library construction. Under different library conversion rate evaluation methods, the library conversion rate of ND610 was significantly better than similar products of Supplier A.
3. Higher effective sequencing depth

The library was constructed using different standards and clinical samples of different qualities as templates. After the sequencing data was normalized, the sequencing depth of the library constructed by ND610 was higher after deduplication.
4. More accurate detection rate of low-frequency mutations

On the 1% FFPE tumor SNV standard, the detection of low-frequency mutation sites in the library constructed by ND610 is consistent with similar products of Supplier A; on the 0.1% lung cancer ctDNA standard, the detection of low-frequency mutation sites in the library constructed by ND610 is better than similar products of Supplier A
Download